
Institute of Molecular Physiology
Junior Principal Investigator
wangzixi@szbl.ac.cn
Timeline
2023 – Present The Institute of Molecular Physiology at Shenzhen Bay Laboratory Junior Principal Investigator
2016-2023 UT Southwestern Medical Center, Postdoctoral Fellow/Instructor
2013-2016 Peking University, Postdoctoral Fellow
2007-2013 Peking University, Ph.D.
2003-2007 Hebei University, B.S.
Current Openings
https://mp.weixin.qq.com/s/13PMtr0Jj2TtJ-al4kUJfQ
Research Areas
My group focuses on identifying new therapeutic targets for fatty liver diseases and liver cancer, the mechanism of liver regeneration, and fundamental biological questions related to liver physiology. The directions include:
1. Identifying new therapeutic targets for NASH;
2. Relief or treatment of cirrhosis and liver cancer;
3. Signaling pathways related to liver regeneration;
4. Molecular basis of zonated metabolism in liver;
5. The role of non-parenchymal cells in liver diseases;
Highlights
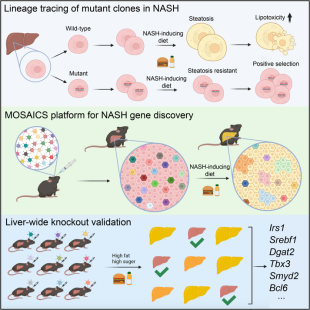
Somatic mutations are common in most individuals, and there is accumulating evidence that the mutation burden increases with age and chronic tissue damage, including fatty liver environment. While the identity and abundance of these mutations are becoming increasingly understood through deep sequencing, fundamental questions about the relevance of these mutations remain unanswered. Many somatic mutations differ from those in cancer samples, and cells that carry them are not fated to become cancerous. To investigate the physiological role of somatic mutations in liver, we established the MOSAICS platform to model widespread somatic mosaicism. MOSAICS screens combined with lineage tracing of mutant hepatocytes support the hypothesis that lipid-laden cells have a fitness disadvantage compared with cells with less fat. This environmental pressure to reduce lipid accumulation fosters the selection of adaptive mutations within NASH livers (Wang, et al. Cell. 2023. See also Research Highlight by Lan, et al. Signal Transduction and Targeted Therapy. 2023). Screens targeting broader genes will be implemented to identify new adaptive mutations for NASH drug discovery.
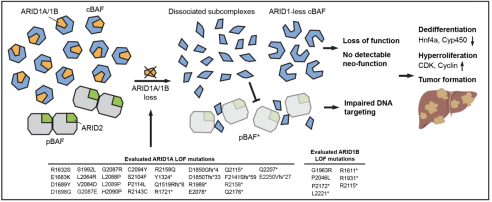
For the recurring oncogenic mutations in liver cancer, we seek strategies explicitly killing cancer cells carrying those mutations. The “synthetic lethal” effect is one such strategy that could be utilized. The principle of synthetic lethality is that when the inactivation of one gene is combined with the loss of another gene, the cell dies, whereas losing an individual gene does not cause noticeable harm to the cell. For cancer cells carrying high-frequency ARID1A mutations, previous in vitro studies have found that ARID1B is its synthetic lethal pair. To verify this widely accepted conclusion on organismal level, we constructed mouse models carrying ARID1A and ARID1B loss-of-function mutations in liver. By contrast, the models showed that dual loss leads to rapid carcinogenesis, indicating that the synthetic lethal effect occurs in a context-dependent manner and suggesting the importance of thoroughly evaluating synthetic lethal strategies on animals (Wang, et al. Nature Cancer, 2020). In subsequent works, we look forward to finding more targeted treatment strategies for different oncogenic mutations.
Honors
2022 Methodological Advances of The Year Award, CRI, UTSW
2013, 2015 Fellowship from Peking-Tsinghua Joint Center for Life Sciences
2004, 2005, 2006 Outstanding Student Scholarship of Hebei University
Selected Publications
1.Wang, Z., Zhu, S., Jia, Y., Wang, Y., Kubota, N., Fujiwara, N., Gordillo, R., Lewis, C., Zhu, M., Sharma, T., Li, L., Zeng, Q., Lin, Y.H., Hsieh, M.H., Gopal, P., Wang, T., Hoare, M., Campbell, P., Hoshida, Y., Zhu, H. Positive selection of somatically mutated clones identifies adaptive pathways in metabolic liver disease. Cell. 186, 1968-1984, (2023)
(Highlighted by Lan, et al. Signal Transduction and Targeted Therapy. 2023)
2.Wang, Z. and Zhu, H. Exploiting liver metabolism for tissue-specific cancer targeting. Nature Cancer. 4, 310-311, (2023) (News & Views)
3.Wang, Z., Chen, K., Jia, Y., Chuang, J-C., Sun, X., Lin, Y-H., Celen, C., Li, L., Huang, F., Liu, X., Castrillon, D.H., Wang, T., Zhu, H. Dual ARID1A/ARID1B loss leads to rapid carcinogenesis and disruptive redistribution of BAF complexes. Nature Cancer. 1, 909–922, (2020)
4.Wei, Y., Wang, Y. G., Jia, Y., Li, L., Yoon, J., Zhang, S., Wang, Z., Zhang, Y., Zhu, M., Sharma, T., Lin, Y. H., Hsieh, M. H., Albrecht, J. H., Le, P. T., Rosen, C. J., Wang, T., and Zhu, H. Liver homeostasis is maintained by midlobular zone 2 hepatocytes. Science. 371 (2021)
5.Huang, F., Huffman, K., Wang, Z., Wang, X., Li, K., Cai, F., Yang, C., Cai, L., Shih, T. S., Zacharias, L. G., Chung, A. S., Yang, Q., Chalishazar, M. D., Ireland, A. S., Stewart, C. A., Cargill, K. R., Girard, L., Liu, Y., Ni, M., Xu, J., Wu, X., Zhu, H., Drapkin, B. J., Byers, L. A., Oliver, T. G., Gazdar, A., Minna, J., and DeBerardinis, R. Guanosine triphosphate links MYC-dependent metabolic and ribosome programs in small cell lung cancer. JCI. 131, 1-18, (2021)
6.Wang, Z., Liu, Y., Huang, S., and Fang, M. TRAF6 interacts with and ubiquitinates PIK3CA to enhance PI3K activation. FEBS Letters. 592, 1882-1892, (2018) (Co-correspondence)
7.Wang, Z., Dang, T., Liu, T., Chen, S., Li, L., Huang, S., and Fang, M. NEDD4L Protein Catalyzes Ubiquitination of PIK3CA Protein and Regulates PI3K-AKT Signaling. JBC. 291, 17467-17477, (2016)
See Google Scholar for full list:
https://scholar.google.com/citations?hl=en&user=UeRzONAAAAAJ&view_op=list_works&sortby=pubdate
Patent
1.Hao Zhu, Zixi Wang, Lin Li. Compositions and methods for treating liver diseases with siRNAs targeting Tbx3. Provisional Patent Application Serial No. 63/328,549; Polsinelli Matter No.: 106546-712251.
2.Hao Zhu, Zixi Wang, Lin Li. Compositions and methods for treating liver diseases with siRNAs targeting Smyd2. Provisional Patent Application Serial No. 63/382,920; Polsinelli Ref. No.: 106546-743413.
3.Hao Zhu, Zixi Wang, Lin Li. Compositions and methods for treating liver diseases with siRNAs targeting Gpam. Provisional Patent Application Serial No. 63/328,593; Polsinelli Matter No.: 106546- 735990.
4.Hao Zhu, Zixi Wang, Lin Li. Compositions and methods for treating liver diseases with siRNAs targeting Cideb. Provisional Patent Application Serial No. 63/328,557; Polsinelli Matter No.: 106546-712249.