2022年5月《生物化学杂志》(JBC)的“酶学”部分,刘玉祥、王涛、崔云凤、李超逸、姜丽芬、饶毅发表论文“生物化学纯化,结合遗传学,揭示STE20磷酸化AMPK相关激酶”。
这篇论文,与饶毅实验室第一篇生物化学论文在同一期JBC上发表。
两篇文章的科学问题都是酶学问题:AMPK及其类似激酶(ARKs),可以被什么激酶所调控。核心方法都是生化分离纯化,辅以分子生物学。第二篇论文还含有质谱分析蛋白质磷酸化位点、基因敲除的细胞和基因敲除的老鼠。
ARKs参与很多重要生物学过程,包括代谢、睡眠、炎症等。
2003年,英国和美国科学家发现LKB1磷酸化ARKs,也就是说LKB1是ARKs的蛋白激酶。
刘玉祥等的第一篇论文通过蛋白质分离纯化发现,MST3可以磷酸化ARKs的SIK3-T221和AMPK-T172,提示ARKs有更多蛋白激酶。
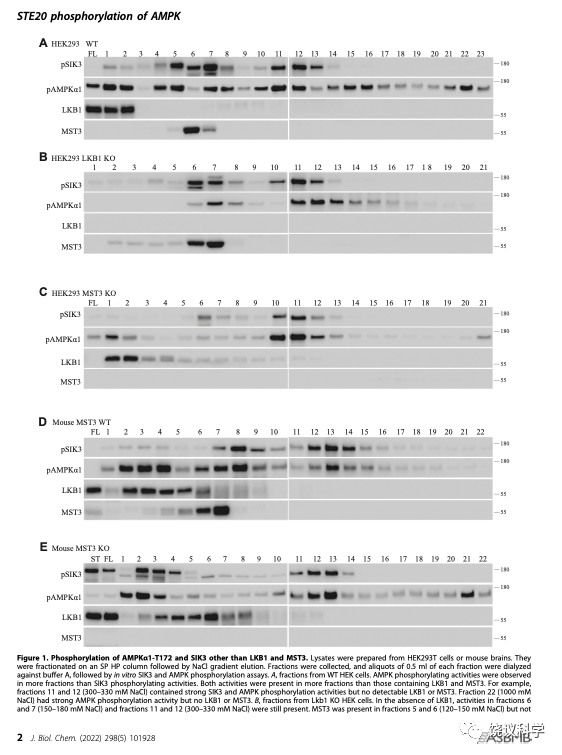
刘玉祥等的第二篇论文起点是,在体外培养的人类细胞HEK中敲除掉MST3之后,还有能够催化SIK3和AMPK的蛋白激酶活性。敲除老鼠的MST3之后,其脑内也仍有能催化SIK3和AMPK的蛋白激酶。通过分析一种色谱柱得到的不同组分,刘玉祥等知道这些活性也不是LKB1。
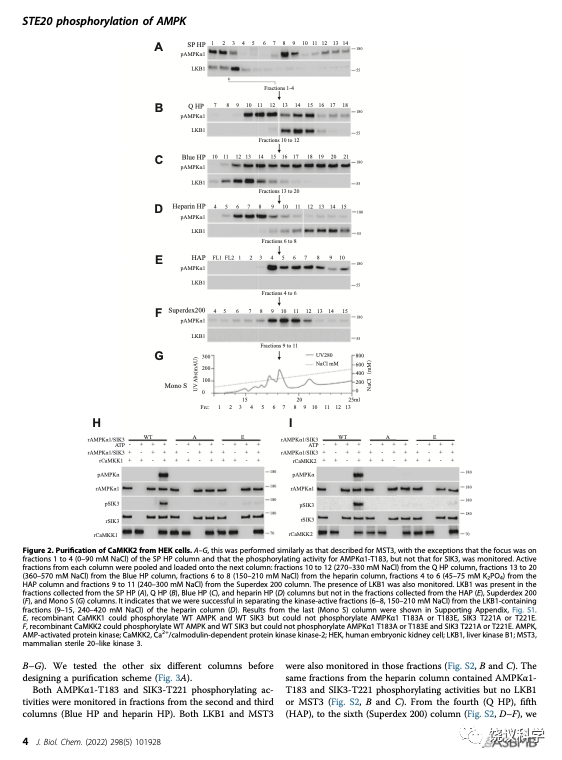
刘玉祥等再通过7根色谱柱,分离纯化的活性组分中含6个激酶,5个为蛋白激酶。其中CAMKK1和2可以催化SIK3-T221和AMPK-T172的磷酸化。CAMKK已经在2005年被发现可以催化AMPK-T172的磷酸化。刘玉祥等再次分离到CAMKK,说明分离纯化的方法是对的,才会重新找到已经发现的。但找到CAMKK不是新发现,只是验证。
刘玉祥等从组分分析发现,应该还有ARKs的更多蛋白激酶。
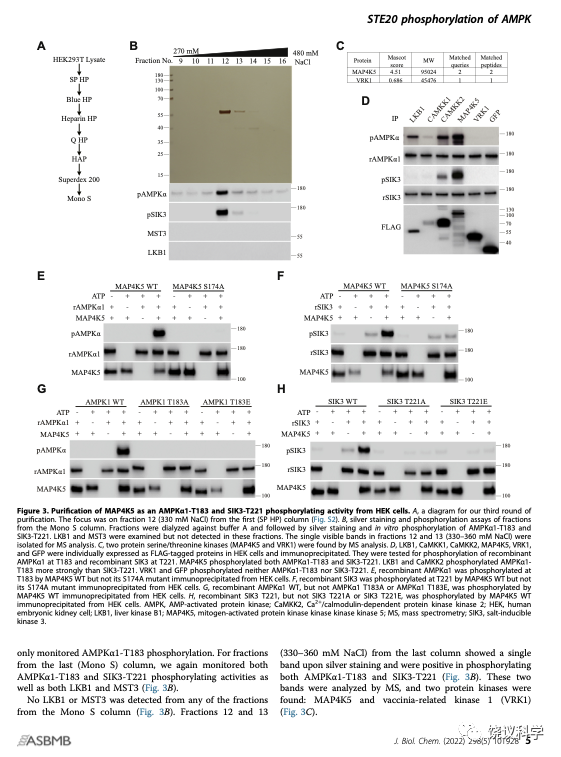
他们用HEK进行第三轮分离纯化。通过7根色谱柱,他们发现活性组分含两个蛋白激酶。其中MAP4K5可以催化SIK3-T221和AMPK-T172的磷酸化。
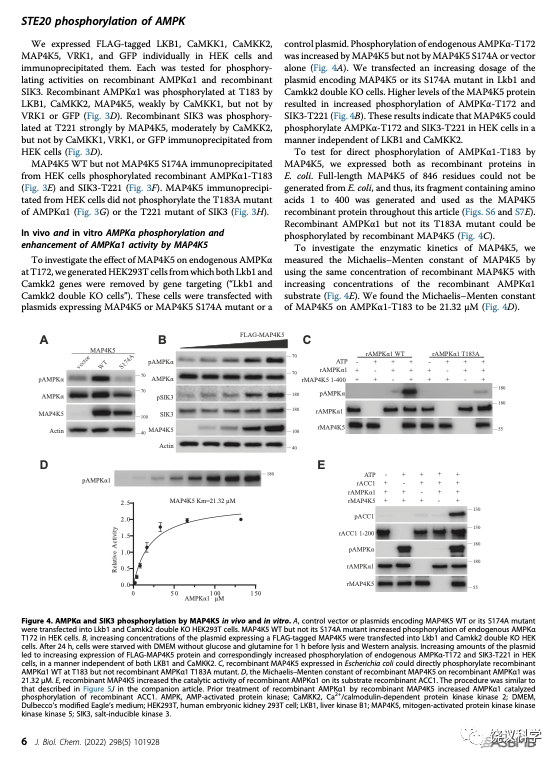
在HEK细胞敲除了LKB1和CAMKK2之后,MAP4K5照样能够催化SIK3-T221和AMPK-T172的磷酸化,说明MAP4K5不需要通过已知的LKB1和CAMKK2来催化SIK3-T221和AMPK-T172的磷酸化。他们进一步的研究验证,细菌表达的MAP4K5也可以有效地催化SIK3-T221和AMPK-T172的磷酸化,说明MAP4K5可以直接催化SIK3-T221和AMPK-T172的磷酸化。
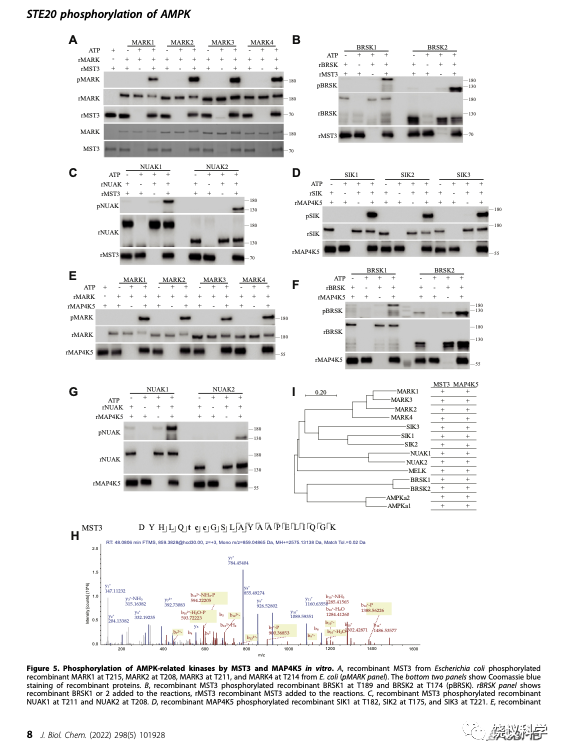
因为已知14个ARKs,刘玉祥、王涛、崔云凤等把14个ARKs的蛋白质都分别由细菌表达,然后作为MST3和MAP4K5的潜在底物,进行生物化学反应。刘玉祥、王涛、崔云凤等发现,MST3和MAP4K5分别都能催化这14个ARKs的磷酸化,包括MARK1-4、NUAK1-2、BRSK1-2、AMPK1-2、SIK1-3、MELK。
非常有趣的是,MST3和MAP4K5都属于哺乳类类似酵母STE20样的蛋白激酶亚家族。这个亚家族共有28个蛋白激酶,包括:MAP4K 1-7、MST 1-5、STK 2/10/39、TAOK 1-3、OSR1、MYO3 A/B、PAK 1-6。
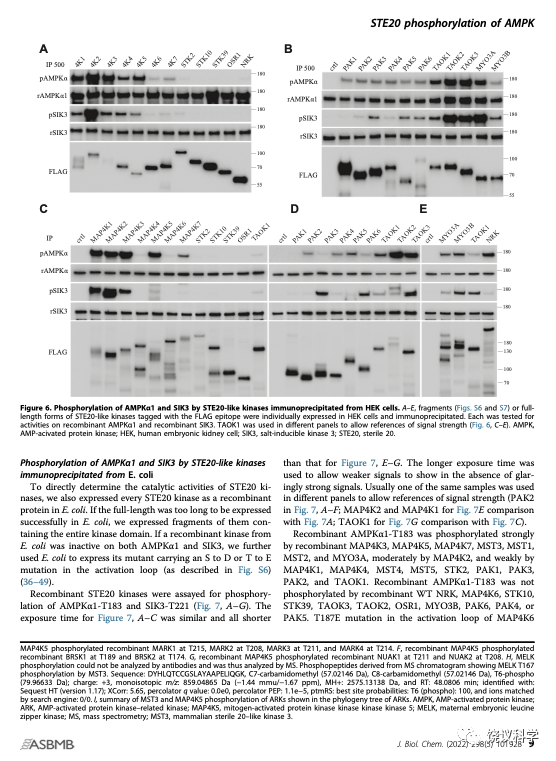
刘玉祥、王涛、崔云凤等在HEK分别表达全部28个STE20蛋白激酶,其中有一个特别难表达,需要培养上百个碟子的细胞,才能拿到足够的量。刘玉祥、王涛、崔云凤等分别检测它们能否催化SIK3-T221和AMPK-T172的磷酸化,结果有些STE20可以,有些STE20不可以。
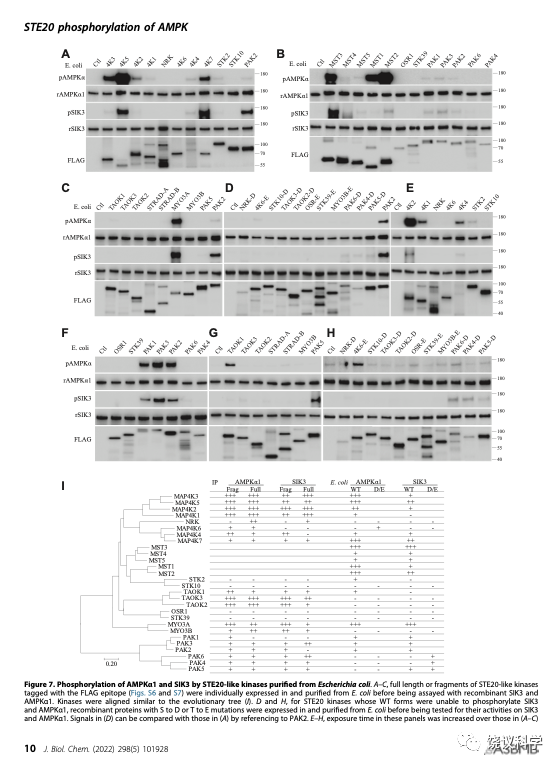
为了避免HEK细胞内其他蛋白质的污染,他们又在细菌分别表达全部28个STE20亚家族蛋白激酶。逐个检测它们能否催化SIK3-T221和AMPK-T172的磷酸化,结果也是有些可以,有些不可以。这些结果大部分与HEK的类似,但不尽相同。
刘玉祥、王涛、崔云凤等突变STE20亚家族蛋白激酶,特别是那些在以上实验看上去没有活性的激酶,看看突变能否提高其活性。有几个的活性可以显现,有些不能因为突变而显现。
刘玉祥、王涛、崔云凤、李超逸、姜丽芬总结了全部28个STE20亚家族蛋白激酶对SIK3-T221和AMPK-T172两个底物的磷酸化作用。
通过三次生物化学分离纯化,通过表达上百个蛋白质及其突变种,刘玉祥、王涛、崔云凤、李超逸、姜丽芬、饶毅等揭示了STE20亚家族蛋白激酶的全新作用,揭示了SIK3-T221和AMPK-T172有多于LKB1、CAMKK等已知蛋白激酶的激酶。
这篇文章,也是生物化学为核心。它揭示了两类蛋白激酶的一种生物化学关系。
这一生物化学关系,是否参与信号转导,是否参与生理学过程,是否被病理学过程所影响或者参与发病,是悬而未决的问题。
解决后面的问题,是一个实验室、还是很多实验室?需要考虑有二十多个酶对至少14个酶。
科学研究取得一个发现可以带来更多问题,需要进一步的研究。
Liu YX, Wang TV, Cui YF, Gao SX and Rao Y (2022). Biochemical purification uncovers mammalian sterile 3 (MST3) as a new protein kinase for multifunctional protein kinases AMPK and SIK3. J Biol Chem 298, 101929.
Liu YX, Wang TV, Cui YF, Li CY, Jiang LF and Rao Y (2022). STE20 phosphorylation of AMPK related kinases revealed by biochemical purifications combined with genetics. J Biol Chem 298, 101928.







Supporting information
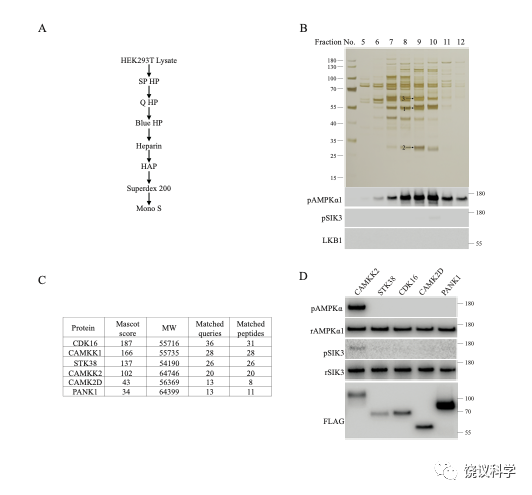
Figure S1
Purification of CaMKK2 from HEK293 T lysates. The scheme is similar to our purification of MST3, with the exception that the focus was on Fractions 1 to 4 (0–90 mM NaCl) from the first SP HP column (shown in Fig. 2). A, A diagram of purification with 7 chromatography columns. B, Silver staining (top panel), and AMPKα1 and SIK3 phosphorylating activities (middle panels) of fractions from the Mono S column. Bottom panel: Western analysis of LKB1. Bands 1, 2 and 3 from Fraction 9 were subjected to MS analysis. C, Kinases detected by MS analysis. CDK16, CaMKK1, STK38, CaMKK2, CaMK2D are protein serine/threonine kinases. PANK1 is pantothenate kinase 1. D, Recombinant AMPKα1 was strongly phosphorylated at T183 by FLAG-tagged CaMKK2, but not by FLAG-tagged CDK16, STK38, CaMK2D or PANK1 immunoprecipitated from HEK cells. Recombinant SIK3 was very weakly phosphorylated at T221 by CaMKK2, but not by CDK16, STK38, CaMK2D or PANK1 immunoprecipitated from HEK cells.
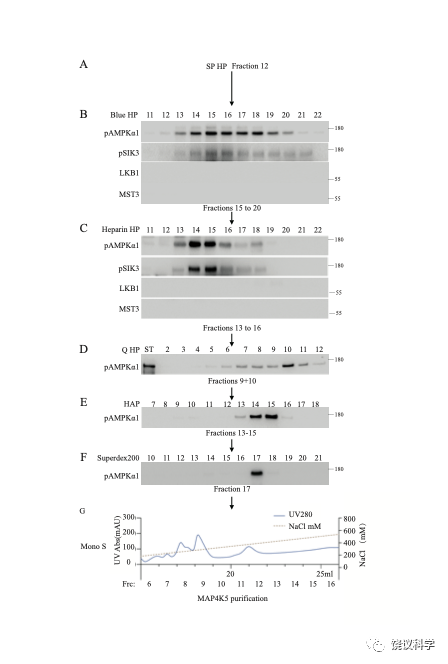
Figure S2
Purification of MAP4K5 from HEK293 T lysates. Purification was similar to that described for MST3, with the exception that, while MST3 was purified from Fraction 7 (180 mM NaCl) of the SP HP column, the present purification was focused on Fraction 12 (330 mM NaCl) from the SP HP column (cf. Fig. 2 of this paper, and Figs. 1 and 2 of the companion article). Results shown here came from purification of 500 mg lysates, passed onto SP HP, Blue HP, heparin HP, Q HP, HAP, Superdex 200 and Mono S columns. AMPKα1 T183 phosphorylating activities were monitored after each fractionation. LKB1 and MST3 were monitored by Western analysis after the Blue HP and the heparin HP columns. Active fractions from each column were pooled and loaded onto the next column: Fractions 15 to 20 (1400–1900 mM NaCl) from the Blue HP column, Fractions 13 to 16 (360–450 mM NaCl) from the heparin HP column, Fractions 9 and 10 (240–270 mM NaCl) from the Q HP column, Fractions 13 to 15 (180–210 mM K2PO4) from the HAP column, and Fraction 17 from the Superdex 200 column. Fractions from Mono S 5/50 Gl column were dialyzed against buffer A and each was assayed for kinase activity and by silver staining. After the initial run, all the rest of the 5000 mg lysates were purified through the same columns, with activities monitoring only after the last column (Mono S). Phosphorylating activities and silver staining of the fractions from Mono S column were shown in Figure 3.
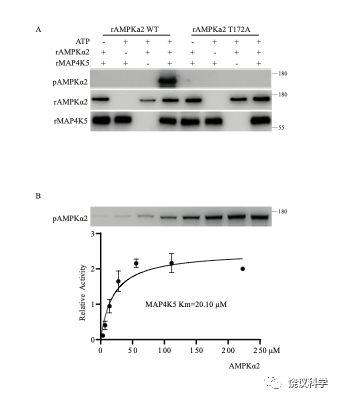
Figure S3
Phosphorylation of AMPKα2-T172 by MAP4K5. A, Recombinant MAP4K5 directly phosphorylated recombinant AMPKα2 WT at T172 in vitro, but not T172A mutant of AMPKα2. B, The Michaelis-Menten constant of recombinant MAP4K5 on AMPKα2 T172 was 20.10 μM.
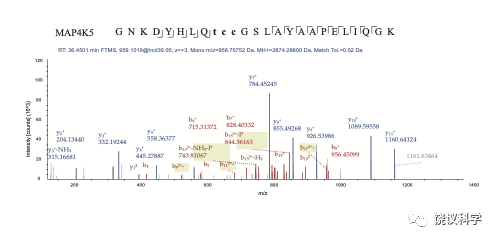
Figure S4
MELK Phosphorylation analyzed by mass spectrometry.Phosphopeptides derived from MS chromatogram showing MELK T167 phosphorylation by MAP4K5. Sequence: GNKDYHLQTCCGSLAYAAPELIQGK, C10-Carbamidomethyl (57.02146 Da), C11-Carbamidomethyl (57.02146 Da), T9-Phospho (79.96633 Da); Charge: +3, Monoisotopic m/z: 958.76752 Da (-2.35 mmu/-2.45 ppm), MH+: 2874.28800 Da, RT: 36.4501 min; Identified with: Sequest HT (v1.17); XCorr:3.56, Percolator q-Value:0.0e0, Percolator PEP:2.9e-6, ptmRS: Best Site Probabilities:T9(Phospho): 100, Ions matched by search engine: 0/0.
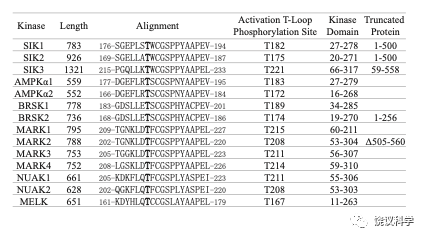
Figure S5
Recombinant AMPK related kinases: fragments and T to E mutants. Attempts to generate full length recombinant proteins in E. coli were successful for some ARKs but not for others (usually the larger ones). Thus, fragments for some ARKs were generated. “Truncated Protein” listed the length of each recombinant protein, and those not listed under “Truncated Protein” were produced as the full-length proteins. The T residue under “Activation Loop Phosphorylation Site” is equivalent to T221 of SIK3 and T172 of AMPKα2.
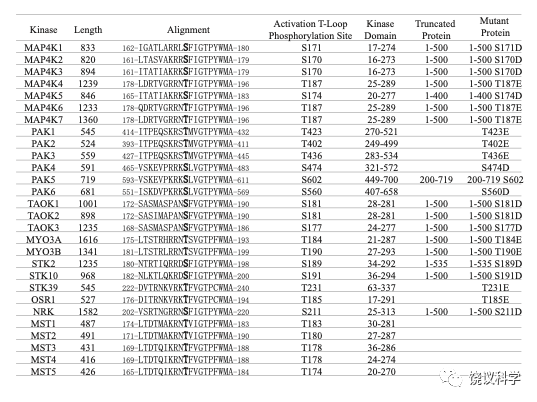
Figure S6
Fragments and S to D or T to E mutants of STE20 kinases. Attempts to generate full length recombinant STE20 proteins in E. coli were successful for some but not for others (usually the larger ones). We thus made fragments (as listed under “Truncated Protein” here). The S or T residue listed under “Activation Loop Phosphorylation Site” could be mutated to D or E for each STE20 kinases (as listed under “Mutant Protein).
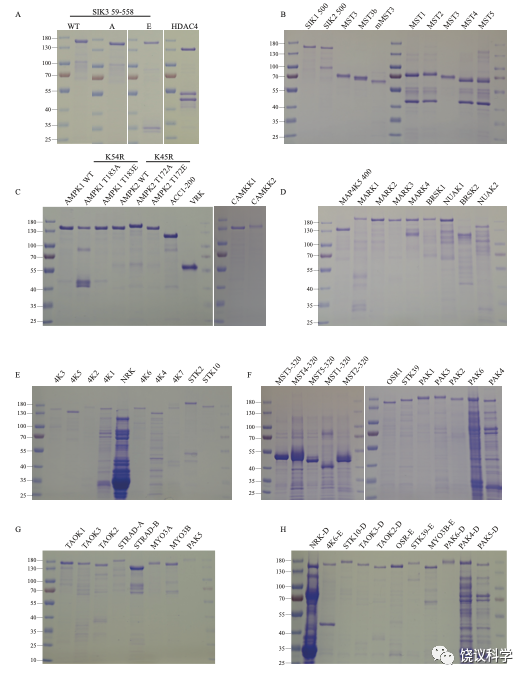
Figure S7
Recombinant proteins expressed in E. coli. Coomassie blue staining of recombinant proteins used in this paper. 1 μg recombinant proteins were denatured by heating at 95 °C with the protein loading buffer and subjected to 12% SDS-PAGE followed by staining with Coomassie blue R250. MST1 to 5, MST3b, mMST3 and VRK1 were made as 3xFLAG tag-kinase-8xHis tag. The others were made as MBP-TEV site-3xFLAG tag-kinase-TEV site-GFP-8xHis tag. TEV site is the cleavage site for Tobacco Etch Virus protease. When these recombinant proteins were used as substrates, they were not cleaved. When they were used as kinases, they were first treated by the TEV protease to release the kinase part from the rest of the fusion protein.